Overview
Welcome to RASP v2.0!
RNA molecules are crucial in numerous biological processes by folding into complex structures. Therefore, studying RNA structure is essential for revealing biological mechanisms and treating human diseases. Since the initial release of the RNA Atlas of Structure Probing (RASP) in 2021, we have actively improved the quantity and quality of RNA structure probing data in the database. We are now launching RASP v2.0, which offers a more comprehensive RNA structure probing dataset with enhanced online analysis capabilities.
Currently, RASP v2.0 supports 438 RNA structure probing datasets covering 85 articles, 24 species, and 39 experimental methods. RASP v2.0 is the first database to integrate footprinting-based and proximity ligation-based structure probing data, facilitating the joint analysis of various RNA structure data. To further improve the data quality of the low structure score coverage data, we used our previously developed models to impute missing structure scores for 59 datasets. RASP v2.0 also provides a variety of advanced structure-related analysis functions, including I) structure score imputation, II) RNA secondary and III) tertiary structure prediction, IV) RBP binding prediction, and. In summary, RASP v2.0 provides a more comprehensive set of RNA structure data through various RNA structure probing methods, facilitating the discovery of higher-order RNA structures and aiding researchers in systematically studying RNA functional mechanisms.
Citation:
RASP v2.0: an updated atlas for RNA structure probing data, Kunting Mu*, Yuhan Fei*,#, Yiran Xu and Qiangfeng Cliff Zhang#, Nucleic Acids Research (2024). [Link to RASP2]
RASP: an atlas of transcriptome-wide RNA secondary structure probing data, Pan Li*, Xiaolin Zhou*, Kui Xu and Qiangfeng Cliff Zhang#, Nucleic Acids Research (2021). [Link to RASP]
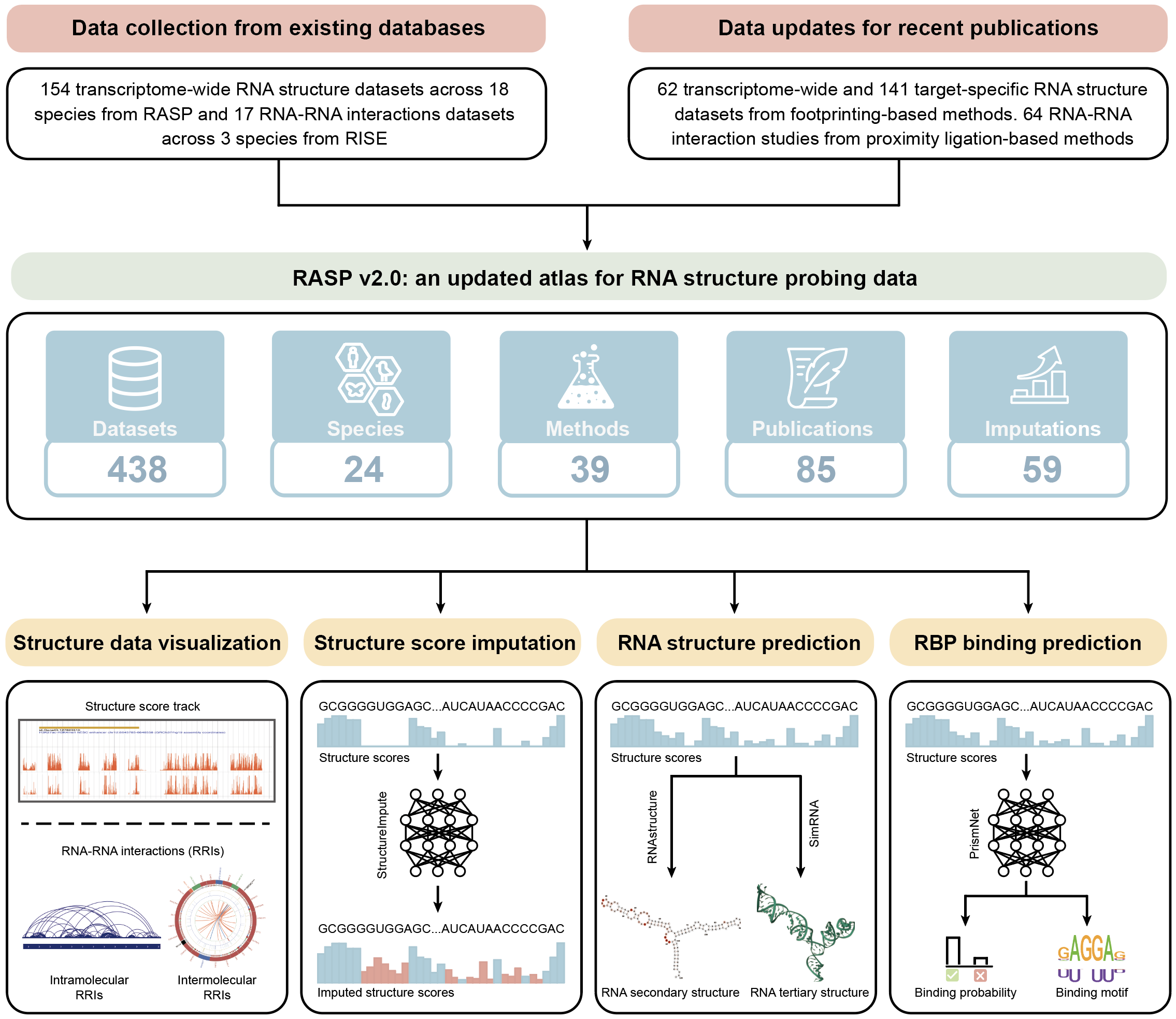
Statistics
What's New?
We updated transcriptome-wide RNA structure probing datasets from 154 to 216 and added 141 target-specific RNA structure probing datasets , increasing the number of species from 18 to 24 and the methods from 18 to 27.
We added 81 RNA-RNA interaction (RRI) datasets from proximity ligation-based structure probing methods, involving 7 species and 12 methods, which were not previously stored in RASP.
We used a deep learning-based model to impute missing structural signals for experimentally undetected regions based on 59 RNA structure probing datasets with low structure score coverage.
We introduced several new features in RASP 2.0 for downstream tasks related to RNA structure, including Structure score imputation, RNA structure prediction, and RBP binding prediction.